baroclinic channel
description
Baroclinic channel tasks implements variants of the Baroclinic Eddies test case from Ilicak et al. (2012).
Polaris includes includes 5 baroclinic channel test cases. All test cases have
at least 2 steps, init, which defines the mesh and initial conditions for the
model, and some variation on forward (given another name in many test cases
to distinguish multiple forward runs), which performs time integration of the
model.
suppported models
These tasks support only MPAS-Ocean.
mesh
The domain is zonally periodic with solid northern and southern boundaries.
The domain size is 160 km in the zonal dimension by 500 km in the meridional
dimension. These dimensions are set in the config file by lx and ly.
Variants of the test case are available at 1-km, 4-km and 10-km horizontal
resolution.
vertical grid
By default, all tests have 20 vertical layers of 50-m uniform thickness. The domain bottom is flat.
# Options related to the vertical grid
[vertical_grid]
# the type of vertical grid
grid_type = uniform
# Number of vertical levels
vert_levels = 20
# Depth of the bottom of the ocean
bottom_depth = 1000.0
# The type of vertical coordinate (e.g. z-level, z-star)
coord_type = z-star
# Whether to use "partial" or "full", or "None" to not alter the topography
partial_cell_type = None
# The minimum fraction of a layer for partial cells
min_pc_fraction = 0.1
initial conditions
Salinity is constant throughout the domain (at 35 PSU). The initial temperature is cooler in the southern half of the domain than in the north, with a gradient between the two halves that is sinusoidally perturbed in the meridional direction.
First the temperature in the depth dimension is defined:
perturb_width, \(T_s\) to
surface_temperature and \(T_b\) to bottom_temperature. The surface
temperature is warmer than at depth. \(z_b\) is the bottom depth.
A fraction is defined to scale the horizontal temperature perturbations, which are uniform with depth:
gradient_width_dist or gradient_width_fraction *
ly. \(y_{mid} = \frac{1}{2} l_y\).
The temperature field is constructed using this fraction of the
temperature_difference, \(\Delta T\), and a fraction of \(\Delta T_{crest}\),
hard-coded as 0.3, applied over a more limited x-extent.
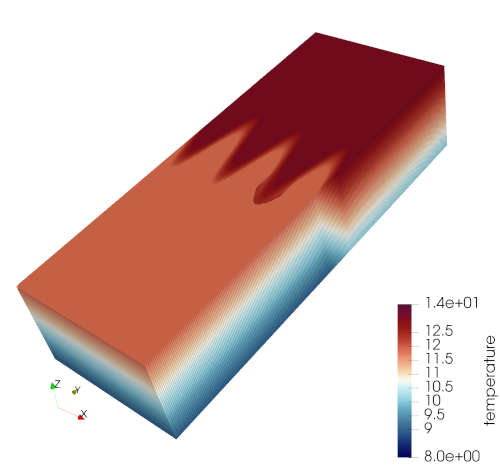
The flow is initially stationary and the Coriolis parameter is uniform across
the domain and defined in the config file by coriolis_parameter.
forcing
N/A
time step and run duration
The time step is determined by the config options dt_per_km so that the time
step is proportional to the resolution. By default, a 10 km-resolution test
has a time step of 5 min. Run duration will be specified for each test case.
Run duration will be discussed for individual test cases.
config options
All 5 test cases share the same set of config options:
# config options for baroclinic channel testcases
[baroclinic_channel]
# the size of the domain in km in the x and y directions
lx = 160.0
ly = 500.0
# time step per resolution (s/km), since dt is proportional to resolution
dt_per_km = 30
# barotropic time step per resolution (s/km), since btr_dt is proportional to
# resolution
btr_dt_per_km = 1.5
# Logical flag that determines if locations of features are defined by distance
# or fractions. False means fractions.
use_distances = False
# Viscosity values to test for rpe test case
viscosities = 1, 5, 10, 20, 200
# Temperature of the surface in the northern half of the domain.
surface_temperature = 13.1
# Temperature of the bottom in the northern half of the domain.
bottom_temperature = 10.1
# Difference in the temperature field between the northern and southern halves
# of the domain.
temperature_difference = 1.2
# Fraction of domain in Y direction the temperature gradient should be linear
# over. Used when use_distances = False.
gradient_width_frac = 0.08
# Width of the temperature gradient around the center sin wave. Default value
# is relative to a 500km domain in Y. Used when use_distances = True.
gradient_width_dist = 40e3
# Salinity of the water in the entire domain.
salinity = 35.0
# Coriolis parameter for entire domain.
coriolis_parameter = -1.2e-4
The default domain size (lx and ly) is designed to be consistent with the
literature, but can be modified by users to suit their needs.
The config options dt_per_km and btr_dt_per_km are used to determine a
time steps that is consistent with a given resolution. Changing these config
options is likely to break the restart, since it assumes a time step of
5 minutes at 10 km, consistent with the default dt_per_km. A user can
safely modify dt_per_km and btr_dt_per_km to control the time step for any
other baroclinic channel tests.
All units are mks, with temperature in degrees Celsius and salinity in PSU.
default
description
ocean/baroclinic_channel/10km/default is the default version of the
baroclinic eddies test case for a short (15 min) test run and validation of
prognostic variables for regression testing.
mesh
See baroclinic channel. Currently, only 10-km horizontal resolution is supported.
vertical grid
See baroclinic channel.
initial conditions
See baroclinic channel.
forcing
See baroclinic channel.
time step and run duration
See baroclinic channel for time step. The run duration is 3 time steps.
config options
See baroclinic channel.
cores
The number of processors is hard-coded to be 4 for this case.
decomp
description
ocean/baroclinic_channel/10km/decomp runs a short (15 min) integration
of the model forward in time on 4 (4proc step) and then on 8 processors
(8proc step) to make sure the resulting prognostic variables are
bit-for-bit identical between the two runs.
mesh
See baroclinic channel. Currently, only 10-km horizontal resolution is supported.
vertical grid
See baroclinic channel.
initial conditions
See baroclinic channel.
forcing
See baroclinic channel.
time step and run duration
See baroclinic channel for time step. The run duration is 3 time steps.
config options
See baroclinic channel.
cores
This test is run on 4 cores for the 4proc step and 8 cores for the 8proc
step.
thread
description
ocean/baroclinic_channel/10km/thread runs a short (15 min) integration
of the model forward in time on 1 threads per processor (1thread step) and
then on 2 threads (2thread step) to make sure the resulting prognostic
variables are bit-for-bit identical between the two runs.
mesh
See baroclinic channel. Currently, only 10-km horizontal resolution is supported.
vertical grid
See baroclinic channel.
initial conditions
See baroclinic channel.
forcing
See baroclinic channel.
time step and run duration
See baroclinic channel for time step. The run duration is 3 time steps.
config options
See baroclinic channel.
cores
The model steps are run on 4 cores. The 1thread step is run on 1 thread
per processor and the 2thread step is run on 2 threads per processor.
restart
description
ocean/baroclinic_channel/10km/restart runs a short (10 min)
integration of the model forward in time (full_run step), saving a restart
file every 5 minutes. Then, a second run (restart_run step) is performed
from the restart file 5 minutes into the simulation and prognostic variables
are compared between the “full” and “restart” runs at minute 10 to make sure
they are bit-for-bit identical.
mesh
See baroclinic channel. Currently, only 10-km horizontal resolution is supported.
vertical grid
See baroclinic channel.
initial conditions
See baroclinic channel.
forcing
See baroclinic channel.
time step and run duration
See baroclinic channel for time step. The full run is two time steps and the restart run is one time step long.
config options
See baroclinic channel.
cores
The number of processors is hard-coded to be 4 for this case.
rpe
ocean/baroclinic_channel/1km/rpe,
ocean/baroclinic_channel/4km/rpe, and
ocean/baroclinic_channel/10km/rpe perform longer (20 day) integration
of the model forward in time at 5 different values of the viscosity (with steps
named rpe_1_nu_1, rpe_2_nu_5, etc.) at any of the 3 supported
horizontal resolutions (1, 4 and 10 km). Results of these tests have been used
to show that MPAS-Ocean has lower spurious dissipation of reference potential
energy (RPE) than POP, MOM and MITgcm models
(Petersen et al. 2015).
mesh
See baroclinic channel.
vertical grid
See baroclinic channel.
initial conditions
See baroclinic channel.
forcing
See baroclinic channel.
time step and run duration
See baroclinic channel for time step. Each run lasts 20 days.
config options
See baroclinic channel. The config option that is specific to this case is:
# Viscosity values to test for rpe test case
viscosities = 1, 5, 10, 20, 200
cores
The number of cores is dynamically computed based on the number of cells. See Model.